Introduction
The similarities and differences among structures of polymorphic molecular compounds have generated considerable commentary. If one can predict the crystal structures for a given molecule, it would help determine the likelihood that different forms exist, and suggest as yet unseen polymorphs of currently known structures.
Crystal Structure Prediction based EA
Since 2012, I have been developing the code to study the crystal packing of small molecules such as ice, ammonia, methane which are of great interests in planetary science. As such, I implemented a method based on the evolutionary algorithms into the USPEX code. In the past years, We have successfully solved a series of elusive crystal structures of a number of molecules which were unable to be fully determined in experiment. In 2015, I also participated the sixth Blind Test for Organic Structure Prediction organized by Cambridge Crystallographic Data Centre (CCDC). Among the five proposed challenges, our team successfully solved the two challenges which are rigid molecule and co-crystal made of nearly rigid molecules. Now we are able to do better with the improved methods. We welcome any collaboration from experiment and theory who has relevant projects in planetary sciences, pharmaceutics, pigments, high energy explosives and organic semiconductors.
See APS Highlights
See ESRF Highlights
New code development
While the previous EA-CSP method has been successfully applied to many molecules. It is getting difficult to maintain the code and implement new functions. Since 2018, my group began to work on a new code PyXtal to provide a more fundamental tool to facilitate the crystal structure prediction.

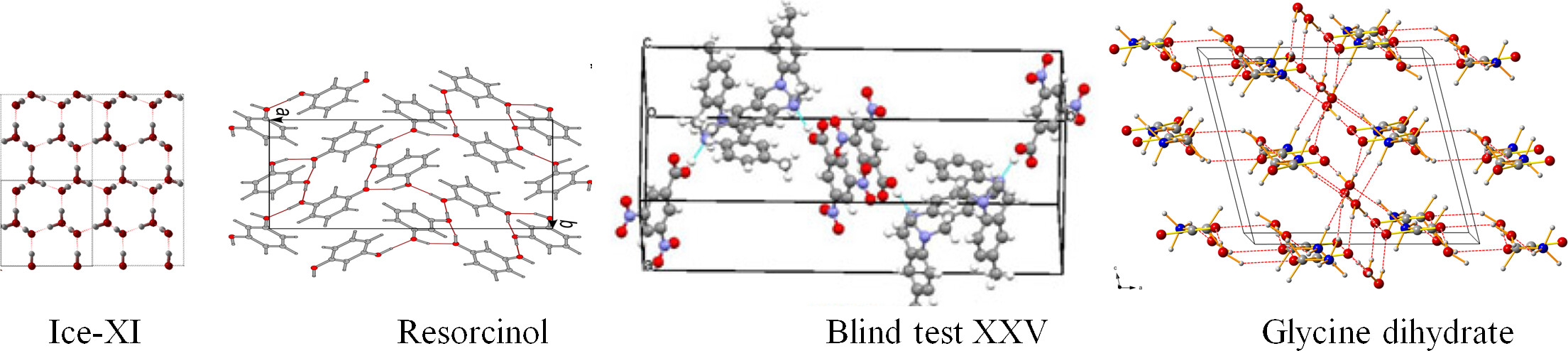
Relevant works
-
Fredericks S., Sayre D., Zhu Q. (2019). PyXtal: a Python Library for Crystal Structure Generation and Symmetry Analysis. (Open Access: PDF)
-
Yang J-X, Zhu, X-L, Hu, C-H, Qiu, M-D, Zhu, Q, Ward, M. D and Kahr B (2019) Inverse Correlation between Lethality and Thermodynamic Stability of Contact Insecticide Polymorphs Cryst. Growth Des.,, 19, 183-1844 (pdf)
-
Tan M, Shtukenberg AG, Zhu SC, Xu WQ, Dooryhee E, Nichols SM, Ward WD, Kahr and Zhu, Q, (2018) ROY revisited, again: the eighth solved structure Faraday Discussion, 211, 477-491 [html]
-
Yang JX, Hu CT, Zhu XL, Zhu Q, et al (2017) DDT Polymorphism and the Lethality of Crystal Forms Angew. Chem. Int. Ed., 56, 10165-10169
-
Shtukenberg AG, Hu CT, Zhu Q, et al (2017) The Third Ambient Aspirin Polymorph Cryst. Growth Des., 17, 3562-3566 (pdf)
-
Shtukenberg AG, Zhu Q, et al (2017) Powder diffraction and crystal structure prediction identify four new coumarin polymorphs Chemical Science, 8 4926-4940 pdf
-
Xu W, Zhu Q, Hu CT (2017). Structure of Glycine Dihydrate: Its implications to Crystallization of Glycine from Solution and Modification of Glycine in Space Angew. Chem. Int. Ed., 129, 2062-2066 pdf APS highlights
-
Zhu Q, Oganov AR, Glass CW, Stokes H.(2012). Constrained evolutionary algorithm for structure prediction of molecular crystals: methodology and applications. Acta. Caryst. B68, 215-226